Researchers develop new way to see single RNA molecules inside living cells
Related images
(click to enlarge)
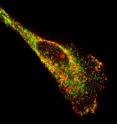
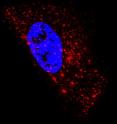
Biomedical engineers have developed a new type of probe that allows them to visualize single ribonucleic acid (RNA) molecules within live cells more easily than existing methods. The tool will help scientists learn more about how RNA operates within living cells. Techniques scientists currently use to image these transporters of genetic information within cells have several drawbacks, including the need for synthetic RNA or a large number of fluorescent molecules. The fluorescent probes developed at the Georgia Institute of Technology circumvent these issues.
"The probes we designed shine bright, are small and easy to assemble, bind rapidly to their targets, and can be imaged for hours. These characteristics make them a great choice for studying the movement and location of RNA inside a single cell and the interaction between RNA and binding proteins," said Philip Santangelo, an assistant professor in the Wallace H. Coulter Department of Biomedical Engineering at Georgia Tech and Emory University.
Details of the probe production process and RNA imaging strategy were published online in the journal Nature Methods on April 6. In addition to Santangelo, Georgia Tech graduate student Aaron Lifland, Emory University associate professor Gary Bassell and Vanderbilt University professor James Crowe Jr. also contributed to this research. This research was funded by new faculty support from Georgia Tech.
In the study, the probes – produced by attaching a few small fluorescent molecules called fluorophores to a modified nucleic acid sequence and combining the sequences with a protein – exhibited single-molecule sensitivity and allowed the researchers to target and follow native RNA and non-engineered viral RNA in living cells.
"The great thing about these probes is that they recognize RNA sequences and bind to them using the same base pairing most people are familiar with in regards to DNA," explained Santangelo. "By adding only a few probes that would bind to a region of RNA, we gained the ability to distinguish a targeted RNA molecule from a single unbound probe because the former lit up two or three times brighter."
For their experiments, the team used a bacterial toxin to transport the probes into living cells – a delivery technique that when combined with the high affinity of the probes for their targets, required significantly fewer probes than existing techniques. The toxin created several tiny holes in the cell membrane that allowed the probes to enter the cell's cytoplasm.
The researchers tested the sensitivity of conventional fluorescence microscopy to image individual probes inside a cell. Previous studies showed that these techniques were able to image an accumulation of probes inside a cell, but the current study demonstrated that individual probes without cellular targets could be observed homogenously distributed in the cytoplasm with no localization or aggregation.
With single-molecule sensitivity accomplished, the researchers investigated whether they could visualize individual RNA molecules using the probes. To do this, they simultaneously delivered probes designed to target a human messenger RNA (mRNA) sequence region and a probe designed with no target in the human genome. They were able to image unbound probes of both types as well as individual RNA molecules that had attached to the former probes.
The imaging technique also allowed the researchers to observe a process called dynamic RNA-protein co-localization, which is the joining of RNA molecules and RNA binding proteins in a single cell.
"We observed substantial transient interactions between proteins and viral RNA molecules that I don't think had ever been seen before with non-engineered RNA," noted Santangelo. "We saw one of the proteins move into a viral RNA granule and reside within it for over a minute before it was released, and we also saw another protein that appeared to dock with a viral RNA granule."
Santangelo is currently trying to improve the probes by making them smaller and brighter, while also using them to investigate viral pathogenesis and other biological phenomena.
"We are excited to use this imaging strategy to study how single viral RNAs travel from the nucleus of a cell to a virus assembly site, how mRNAs are regulated by location and time, and RNA trafficking in neurons," added Santangelo.
Source: Georgia Institute of Technology Research News
Other sources
- Researchers develop new way to see single RNA molecules inside living cellsfrom Biology News NetMon, 6 Apr 2009, 23:21:16 UTC
- Seeing Single RNA Molecules Inside Living Cells: Researchers Develop New Methodfrom Science DailyMon, 6 Apr 2009, 18:28:19 UTC
- Researchers develop new way to see single RNA molecules inside living cellsfrom PhysorgMon, 6 Apr 2009, 18:21:13 UTC
- Researchers Develop New Way to See Single RNA Molecules in Living Cellsfrom Newswise - ScinewsMon, 6 Apr 2009, 17:42:09 UTC