Scientists characterize protein essential to survival of malaria parasite
A biology lab at Washington University has just cracked the structure and function of a protein that plays a key role in the life of a parasite that killed 655,000 people in 2010. The protein is an enzyme that Plasmodium falciparum, the protozoan that causes the most lethal form of malaria, uses to make cell membrane.
The protozoan cannot survive without this enzyme, but even though the enzyme has many lookalikes in other organisms, people do not make it. Together these characteristics make the enzyme an ideal target for new antimalarial drugs.
The research was published in the January 6 issue of the Journal of Biological Chemistry (JBC) as "Paper of the Week" for that issue.
The work also will be featured in ASBMB Today (the newsletter of the American Society for Biological Molecular Biology, which publishes JBC).
Sweating the cold room
The protein's structure might have remained an enigma, had it not been the "unreasonable optimism" of Joseph Jez, PhD, associate professor of biology in Arts & Sciences, which carried his team through a six-year-long obstacle course of failures and setbacks.
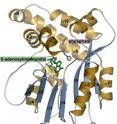
"What my lab does is crystallize proteins so that we can see what they look like in three dimensions," Jez says. "The idea is that if we know a protein's structure, it will be easier to design chemicals that would target the protein's active site and shut it down," Jez says.
The lastest discovery is the culmination of a project that began years before when Jez was working at the Danforth Plant Science Center in St. Louis and collaborating with scientists at the local biotech startup Divergence. "At the time, C. elegans had just been sequenced and the Divergence scientists were looking at using it as an easy model to work out the biochemistry of parasitic nematodes," Jez says.
C. elegans is a free-living nematode, or microscopic roundworm, but many nematodes are parasitic and cause disease in plants, livestock and people.
During this project, Lavanya Palavalli, a summer intern working with Jez, crystallized the C. elegans version of the enzyme. The job of the enzyme, phosphoethanolamine methyltransferase, thankfully abbreviated to PMT, is to add methyl groups to a starting molecule, phosophoethanolamine.
"When Soon Goo Lee later took up the project," says Jez, "the plan was to try to grow better crystals of the C. elegans protein, ones good enough to get readable X-ray diffraction patterns.
Two years later, the crystals were looking better but still not good enough.
So Jez suggested that Lee go after homologous (look-alike) proteins in other organisms. "Even though the proteins are homologous, each has a different amino acid sequence and so will behave differently in the crystallizations," Jez says. "Lee went from working with two C. elegans proteins to three plant proteins, two other nematode proteins and then the Plasmodium protein," Jez says.
"He took all six of those PMT versions into the crystallization trials to maximize his odds," Jez says.
"To crystallize a protein," Jez says, "we put a solution of a salt or something else that might work as a desiccant in the bottom of a small well. And then we put a drop of our liquid protein on a microscope cover slip and flip it over the top of the well, so the drop of protein is hanging upside down in the well."
"What we're trying to do is to slowly withdraw water from the protein. It's exactly like making rock candy, only in that case, the string hanging into the jar of sugar solution helps to withdraw water," he says.
The difference is that sugar wants to form crystals and proteins are reluctant to do so.
"There are 24 wells to a tray, and we usually screen 500 wells per protein at first," Jez says. "Lee had eight proteins and so his first pass was to screen 4,000 conditions. And then he had to try different combinations of ligands to the proteins and crystallize those. This is why it took a few years to finally get where he needed to go."
Road trip! The scientists need crystals -- preferably nice, big ones -- to stick in the path of an X-ray beam at Argonne National Laboratory in Chicago. (If the crystal is a good one, and all the atoms are lined up in a repeating array, the scattered X-rays will produce a clear pattern of spots.)
Embedded in that pattern is the mathematical information needed to back-calculate to the position of the atoms in the protein, a process a bit like throwing a handful of pebbles in a lake and then calculating where they landed by the pattern of waves arriving at the shoreline.
Lee got the PMT from Haemonchus contortus to crystallize first, but there were technical issues with the diffraction pattern that would have made solving it technically and computationally very demanding.
"When the Plasmodium enzyme finally crystallized, Soon got four crystals kind of stacked on top of each other and each of them was paper thin," Jez says.
"I never thought it would work, but we took them to Argonne anyway and he actually did surgery under the microscope and cracked off a little tiny piece of it."
To everyone's surprise, he got a clean diffraction pattern from the crystal. "Because the Plasmodium enzyme was the smallest one and the easiest to work on, we pushed that one first," Jez says.
The moment of truth "Once we had a Plasmodium crystal that was diffracting really well, we could try back-calculating to see whether we could extract the atom positions from the data," Jez says.
After the computer finished its calculations, Lee clicked a mouse button to see the results, which would reveal whether his years of work finally would pay off.
When Lee clicked the mouse, he got an electron density map in exceptionally sharp focus.
"When you see a map like that, it's like suddenly the wind has kicked up and you're sailing free," Jez says, "because there's this moment, like, before you click that button, no one has ever seen how this protein is put together in three dimensions. You're the first person to ever see it.
"The irony of it is we got such good quality diffraction pattern and electron density maps off such an ugly crystal," he says.
Lock and load "Once you have the electron density map, the task is to build a structure that matches the amino acid sequence of the protein," Jez says.
"The first thing you do is put in the amino acid backbones and connect them together to form a chain. It's like having a long thread, each inch of which is an amino acid, and your job is to take that thread and move it in three dimensions through that electron density map."
The next step is to add the side chains that make one amino acid different from another, Jez says. "The amino acid sequence is known," he says. "Your goal is to match the way you string together the amino acids in the electron density map to that sequence."
"Once you have the overall structure, you can start to figure out how the enzyme works. The PMT enzyme is trying to join two molecules," Jez says. "To do that, it has to lock them in place so that the chemistry can happen, and then it has to let go of them.
"We think the protein has a lid that opens and closes," he says. "The active site stays open until the substrates enter, and then the lid clamps down, and when it clamps down it actually puts the substrates together."
Calling Bill and Melinda Gates Not only do infections by Plasmodium falciparum cause the most severe form of malaria, about 40 percent of the human population lives in areas where the parasite is endemic. Moreover, drugs that used to be effective against malaria are beginning to fail, in part because widespread drug counterfeiting has led to resistance.
New anti-malarial drugs are desperately needed, and the PMT protein is an ideal target. If PMT is disabled, the protozoan can't make cell membranes and it dies. Moreover, a drug that would kill Plasmodium might have minimal side effects on patients.
Although the process of identifying compounds that would target PMT is in the early stages, a handful of anti-parasitical compounds used to treat diseases are known to block PMT as well.
As for Lee, he has had a hard go of it, but now things are breaking his way. Plasmodium PMT is giving up its secrets, and the plant and nematode PMTs are coming along as well.
When he clicked the mouse button and a clean electron density map came up, he says, it was like seeing "the light at the end of a five-year-long tunnel."
Source: Washington University in St. Louis
Other sources
- Scientists characterize protein essential to survival of malaria parasitefrom Biology News NetMon, 9 Jan 2012, 1:00:28 UTC
- Scientists characterize protein essential to survival of malaria parasitefrom Science DailySat, 7 Jan 2012, 21:30:48 UTC
- Scientists characterize protein essential to survival of malaria parasitefrom PhysorgSat, 7 Jan 2012, 0:00:43 UTC