Activating ALC1: With a little help from friends
Chromatin remodeling -- the packaging and unpackaging of genomic DNA and its associated proteins -- regulates a host of fundamental cellular processes including gene transcription, DNA repair, programmed cell death as well as cell fate. In their latest study, scientists at the Stowers Institute for Medical Research are continuing to unravel the finicky details of how these architectural alterations are controlled. Through a series of biochemical experiments, Stowers Investigators Ron Conaway, Ph.D., and Joan Conaway, Ph.D., and their team discovered that chromatin remodeling enzyme and suspected oncogene ALC1 (short for Amplified in Liver Cancer 1) is activated through an unusual mechanism: Its shape shifts in the presence of its activators. Their finding identifies a new instrument in cells' molecular repertoire of chromatin-remodeling tools and a potential cancer therapeutic target.
One of the main tasks of chromatin remodeling enzymes, Ron Conaway explains, is "to make DNA accessible so events like repair and gene transcription can occur." Postdoctoral research associate and first author Aaron Gottschalk, Ph.D., previously figured out that ALC1 required protein partners to activate its remodeling function. Publishing in an upcoming issue of the Journal of Biological Chemistry, he dissects the mechanism by which this occurs.
ALC1 and its ilk have a common protein domain, SNF2, that uses the energy of ATP hydrolysis to move nucleosomes -- the basic repeating units of chromatin -- around, in a process called nucleosome sliding.
"I was intrigued because ALC1 has a unique macrodomain not found on any other SNF2 family member," Gottschalk says. His interest was further piqued when he found that while most ATP-dependent chromatin remodelers function as large multi-protein complexes, ALC1 appeared to work by itself. At the same time, where most of its group-happy family members readily demonstrated nucleosome sliding activity in vitro, ALC1 was not only a lone ranger but also "completely dead on its own."
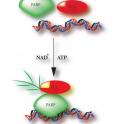
Gottschalk deduced that ALC1 may function independently, but it needs a boost from a couple of sidekicks: PARP1, an enzyme that responds to several kinds of DNA damage; and NAD+, the substrate by which PARP1 transfers chains of poly (ADP-ribose) onto itself and other target proteins, in a process called PARylation.
Only when PARP1 and NAD+ are on the scene does ALC1 spring into action, altering the accessibility of DNA by shifting nucleosomes around. Gottschalk's earlier findings were published in the Proceedings of the National Academy of Sciences in June 2009.
"We then extended this research," Conaway says, "and the upshot of our recent JBC paper is that ALC1 is likely activated through a series of physical interactions. ALC1's unique macrodomain can bind PAR, and protein-protein interactions also occur between ALC1 and PARP1." Gottschalk and coauthor Rushi Trivedi, a graduate student in the Biochemistry & Molecular Biology department at KU Medical Center, developed a novel footprinting assay that enabled this observation. Rather than merely activating ALC1 and moving on, the researchers found that the trio of PARP1, NAD+ and ALC1 hangs out in a stable complex.
"So PARylated PARP1 and NAD+ are allosteric effectors -- by binding to ALC1, they alter its state from dormant to active," Conaway says. "It's an interesting mechanism that's different from how most other chromatin remodelers work. It may also help explain other evidence that PARP1 has the ability to rearrange nucleosomes and reorganize chromatin; this could be one way by which PARP1 exerts its influence."
Apart from its role in modifying chromatin structure, not much is currently known about ALC1. It's regarded as a possible oncogene, being found in excess in hepatocellular carcinoma cells and because overexpression of ALC1 in mice induces spontaneous tumors.
PARP1, on the other hand, has attracted plenty of interest as a potential anticancer drug target, due to its importance in maintaining genomic integrity. For example, in breast cancer cells lacking BRCA1 or BRCA2 function, blocking PARP could effectively remove the cells' last line of defense against DNA-damaging chemotherapy agents. To date, no PARP inhibitor has made it past phase III clinical trials, but pharmaceutical companies continue to chip away at the challenges around optimizing this form of targeted cancer therapy.
"A better understanding of the in-depth biochemistry we're uncovering on ALC1 and PARP1 may, in the long term, ultimately lead to new or more refined therapeutic strategies," Gottschalk says.
Meanwhile, having observed that ALC1 comes to life upon interacting with friends, Gottschalk now wants to understand precisely how this activation happens. With a knockout ALC1 mouse model handy, plans are afoot to extend his studies to an in vivo characterization of a chromatin remodeler that boasts an illustrious family pedigree, yet stands apart in the crowd.
Funding for this study came from a National Institute of General Medical Sciences grant (GM41628), and a grant to the Stowers Institute from the Helen Nelson Medical Research Fund at the Greater Kansas City Community Foundation.
Source: Stowers Institute for Medical Research
Other sources
- Chromatin remodeling: Activating ACL1 with a little help from 'friends'from Science DailyThu, 29 Nov 2012, 21:31:25 UTC
- Activating ALC1: With a little help from friendsfrom PhysorgThu, 29 Nov 2012, 21:00:42 UTC
- Activating ALC1: With a Little Help From Friendsfrom Newswise - ScinewsThu, 29 Nov 2012, 20:31:18 UTC