Molecule serves as a key in some protein interactions
Research led by St. Jude Children's Research Hospital scientists has identified an unexpected mechanism facilitating some protein interactions that are the workhorses of cells and, in the process, identified a potential new cancer drug development target. The discovery involves a chemical known as an acetyl group. An estimated 85 percent of human proteins have this chemical added to the amino acid at one end of the protein. The addition comes in a process known as N-terminal acetylation. N-terminal acetylation occurs shortly after proteins are assembled. Although it has long been known that proteins are N-terminally acetylated, until now it was unknown how such acetylation could serve specific functions.
The findings came from scientists studying a system cells use to regulate the fate and function of proteins. The researchers showed that much like a key must fit precisely to work a lock, the acetylated end of one enzyme fits perfectly into a deep pocket on the surface of another protein. The connection helps accelerate the activity of a protein complex that is involved in regulating cell division and that has been linked to cancer. The research appears in the Nov. 4 print edition of the journal Science.
The findings have potential implications for drug discovery and for understanding basic mechanisms governing the interaction of possibly thousands of proteins, said the study's senior author, Brenda Schulman, Ph.D., a member of the St. Jude Department of Structural Biology and a Howard Hughes Medical Institute investigator.
"The work presents a major new concept in protein-protein interactions," she said. "This raises the question of whether similar 'keys' on thousands of different proteins also unlock doors to allow them to function."
The research offers the first view of how N-terminal acetylation mediates protein interactions, bringing proteins together to do the work of cells. "These findings raise the possibility that N-terminal acetylation plays an important role in the function of a large number of proteins in the cell," said first author, Daniel Scott, Ph.D. a scientist in Schulman's laboratory.
The pocket where N-terminally acetylated proteins bind may also be a good target for small molecules designed to block protein interactions that lead to many diseases, including cancers, Schulman said.
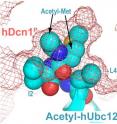
Schulman and her colleagues discovered the pivotal role N-terminal acetylation plays while studying interactions between the proteins Ubc12 and Dcn1. Other researchers have identified human Dcn1 as an oncogene that promotes some squamous cell head, neck and lung cancers.
The focus of the current study was the role the enzymes played in regulating activity of another complex, known as cullin-RING. The cullin-RING complex is the command center of a tagging system that cells use to modify a protein's function or to mark a protein for degradation. The cullin-RING targeted proteins include those that control such important biological processes as cell division and the immune response.
Dcn1 is bound to the cullin protein. In previous studies, Schulman and her colleagues showed that in yeast the Ubc12 and Dcn1 interaction led Ubc12 to transfer its cargo, a protein called NEDD8, to cullin. That step dramatically accelerated the activity of cullin-RING.
But a major question was how human Dcn1 and Ubc12 interact. The breakthrough came when the researchers realized that Ubc12 is among the 35 to 50 percent of proteins in which methionine is the amino acid involved in N-terminal acetylation. The investigators used a variety of laboratory techniques to demonstrate that the acetylated methionine of Ubc12 was essential to the Ubc12-Dcn1 interaction. Evidence included an X-ray image that shows Ubc12's acetyl-methionine buried in a pocket on the surface of Dcn1.
"The size and shape of the pocket indicate it might be a completely new route to generating small- molecule inhibitors," Schulman said. Pharmaceutical companies are already targeting other parts of the same NEDD8 pathway, and one experimental drug has entered clinical trials.
Not only does Ubc12's acetylated tail fit perfectly into the pocket, but acetylation solved another problem that would have made the interaction of Ubc12 and Dcn1 difficult. Without acetylation, Ubc12 and the Dcn1 pocket would repel each other much like oil and water. Acetylation neutralizes the charge on Ubc12, allowing the interaction to occur, Schulman said.
Other authors are Julie Monda of St. Jude; J. Wade Harper of Harvard Medical School; and Eric Bennett, formerly of Harvard Medical School and now at University of California, San Diego.
The research was funded in part by the National Institutes of Health, Howard Hughes Medical Institute, Millennium Pharmaceuticals, Damon Runyon Cancer Research Foundation and ALSAC.
Source: St. Jude Children's Research Hospital
Other sources
- Molecule serves as a key in some protein interactionsfrom Science DailyMon, 7 Nov 2011, 12:30:22 UTC
- Molecule Serves as a Key in Some Protein Interactionsfrom Newswise - ScinewsFri, 4 Nov 2011, 16:01:37 UTC
- Molecule serves as a key in some protein interactionsfrom PhysorgFri, 4 Nov 2011, 15:00:39 UTC