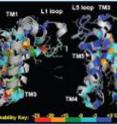
Is it a rock, or is it Jell-O? Defining the architecture of rhomboid enzymes
Johns Hopkins scientists have decoded for the first time the "stability blueprint" of an enzyme that resides in a cell's membrane, mapping which parts of the enzyme are important for its shape and function. These studies, published in advance online on June 14 in Structure and on July 15 in Nature Chemical Biology, could eventually lead to the development of drugs to treat malaria and other parasitic diseases. "[It's] the first time we really understand the architectural logic behind the structure of the enzyme," says Sinisa Urban, Ph.D., an associate professor of molecular biology and genetics at the Johns Hopkins University School of Medicine and an investigator at the Howard Hughes Medical Institute, who with his team has unlocked the mysteries of a special class of enzymes called rhomboid proteases.
Rhomboid proteases are present in many different organisms, and are a unique type of enzyme that resides in the cell's membrane where they cut proteins. Previously Urban and his colleagues demonstrated that the rhomboid enzyme is critical for Plasmodium falciparum, the parasite that causes malaria, to successfully invade red blood cells, a step that ultimately leads to infection. Urban says understanding the stability of rhomboid protease shape may impact the design of enzyme inhibitors -- potential drugs. "These enzymes have no selective inhibitors," says Urban. "We really need to understand how [the enzyme] works -- is it as stiff as a rock, or is it more gummy, like Jell-O?"
One challenge of studying rhomboid enzymes is that they are surrounded by membranes, making them more difficult to manipulate and work with. To address this, Urban's research team turned to a technique known as thermal light scattering, which heats enzyme samples to progressively higher temperatures while measuring the amount of light bouncing back off of the molecules. Enzymes that have broken from their normal shape will scatter light differently, and the temperature at which this occurs (in effect, the breaking point of the enzyme) indicates the inherent stability of the enzyme.
The researchers first precisely measured the stability of the rhomboid enzyme from E. coli bacteria. Surprisingly, says Urban, the rhomboid enzyme was more "Jell-O-like" than other membrane proteins with similar shapes. He guesses that this "jiggly shape" may help rhomboid proteases interact with other proteins that it cuts. To find which parts of the enzyme are most important for maintaining shape and which parts are more crucial for function, the researchers then made and tested 150 differently altered versions of the enzyme. They found four main regions important for maintaining shape and at least two regions important for function.
The researchers also took advantage of computer simulations to test their ideas about how the enzyme functions. Using a computer program model of the enzyme, they programmed in features of its natural membrane environment, which consists mostly of fats and is very limited in water. The computer program then simulated how this environment might influence the enzyme. Researchers found that the enzyme contains a special internal pocket for holding water molecules -- a great advantage in its natural, water-limiting environment.
"We're very excited about our findings and are especially curious about the versions of the enzyme that lost function despite no obvious change in stability or shape," says Urban. Ultimately he hopes that a better understanding of rhomboid proteases will lead to new therapies for treating malaria and other parasitic diseases.
These studies were supported by the Howard Hughes Medical Institute, a National Institute of General Medical Sciences grant (GM079223), a National Institute of Allergy and Infectious Diseases grant (AI066025), the National Science Foundation (NSF) and the David and Lucile Packard Foundation.
Other researchers who participated in this study include Rosanna Baker, Yanzi Zhou, Syed Moin and Yingkai Zhang.
Source: Johns Hopkins Medicine
Other sources
- Is it a rock, or is it Jello? Defining the architecture of rhomboid enzymesfrom Science DailyTue, 31 Jul 2012, 16:50:23 UTC
- Is it A rock, or is it Jell-O? Defining the architecture of rhomboid enzymesfrom PhysorgTue, 31 Jul 2012, 13:31:06 UTC