Scientists make stunning inner space observations
Related images
(click to enlarge)
Scientists using high-powered microscopes have made a stunning observation of the architecture within a cell -- and identified for the first time how the architecture changes during the formation of gametes, also known as sex cells, in order to successfully complete the process. The findings by the international team led by the University of Leicester could impact on the treatment of disorders caused by a misregulation of cellular structures called microtubules. These disorders include Down's Syndrome, lissencephaly (a brain formation disorder) or cancer.
Researching yeast cells, the scientists expect that the information obtained will bring novel insights into the basic mechanisms of cellular regulation, which may be applicable to higher organisms including humans.
The study led by Dr. Kayoko Tanaka of the Department of Biochemistry focused on microtubules -- fibrous structures that play essential roles in a cell. Dr Tanaka had a BBSRC New Investigator award between 2008-2011 to conduct this research which is published in Current Biology.
By exploiting yeast cells, the researchers have discovered for the first time the precise structure adopted by microtubules, which play a vital role in the process of gamete formation, and identified the protein responsible for creating the structure. They further found that the protein needs to be regulated in order to complete gamete formation, failure of which may lead to production of gametes with the wrong number of chromosomes. In humans, these may contribute to disorders such as Down's Syndrome that result from chromosomal abnormalities.
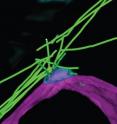
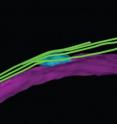
With the use of high-powered fluorescent and confocal microscopes at the University of Leicester, as well as electron microscopes at the European Molecular Biology Laboratory Heidelberg, Dr Tanaka and her team were for the first time able to 'visualise' the architecture of the microtubules in the yeast cell undergoing gamete formation.
Dr Tanaka said: "Microtubules have attracted much attention because they are such a fundamental structure in the cell to direct numbers of dynamic cellular activities. For example, they play an essential role in chromosome segregation during gamete formation. Also, it is known that mutations in the proteins that modulate microtubule dynamics can result in lissencephaly. Furthermore, empirically, microtubule-targeted drugs that disrupt the microtubule dynamics have been shown to be effective in cancer treatment.
"When cells form gametes or various neuronal cells, microtubule reorganisation occurs to help the transition. We used yeast cells to investigate this reorganisation of the microtubule.
"In collaboration with Dr Claude Antony's Group at EMBL (Heidelberg), leading experts in the electron microscopy and tomography, we, for the first time, could have a very close look at the microtubule arrangement in yeast cells undergoing sexual differentiation using EM tomography. Dr Charlotta Funaya in Antony's group has done the amazing EM work that requires extreme accuracy and dedication.
"We found that a radial microtubule array emanating from a site called microtubule organising centre (MTOC) was readily recognisable. Strikingly however, the structure of the MTOC during early sexual differentiation is distinctively different from the well-known one from proliferating cells. We named this novel structure the radial microtubule organising centre (rMTOC).
"I am really excited by this discovery as it has been my aspiration to employ electron tomography to look at the cell structure during the yeast sexual differentiation process, where amazing reorganisation of microtubule architecture takes place. We were extremely lucky to be able to collaborate with Dr Antony's group in EMBL Heidelberg. We were stunned when we saw the striking rMTOC structure."
In another advance, the team collaborated with the Biological Mass Spectrometry Facility led by Dr Duncan Smith at the Paterson Institute for Cancer Research (Manchester) to investigate a protein, called Hrs1/Mcp6, responsible for the development of the rMTOC structure.
Dr Tanaka added: "In addition, collaboration with Dr Smith and his colleagues was fantastic, without their help, it would not have been possible to prove all the modifications of the Hrs1/Mcp6 molecule.
"Taken together with cell biological analyses done by Dr Shivanthi Samarasinghe in my lab, we reveal that the dynamic microtubule rearrangement involves transient generation of a specialized structure, rMTOC, which is later subjected to a rigorous down-regulation to support a smooth transition to the next stage. As Hrs1/Mcp6 has likely mammalian counterparts, this paradigm of rigorously controlled, transient, specialized structures to organise micorutuble architecture may be conserved across species.
"The storyline, or our hypothesis, of our work existed more than 5 years ago when I came to Leicester to start my own lab for the first time. I have always wanted to prove it... and it is almost surreal to finally obtain substantial evidences that support our hypothesis."
Source: University of Leicester
Other sources
- High-powered microscopes reveal inner workings of sex cellsfrom Science DailyTue, 1 May 2012, 15:30:35 UTC
- Scientists make stunning inner space observationsfrom PhysorgTue, 1 May 2012, 15:00:35 UTC