Penn research makes advance in nanotech gene sequencing technique
The allure of personalized medicine has made new, more efficient ways of sequencing genes a top research priority. One promising technique involves reading DNA bases using changes in electrical current as they are threaded through a nanoscopic hole. Now, a team led by University of Pennsylvania physicists has used solid-state nanopores to differentiate single-stranded DNA molecules containing sequences of a single repeating base.
The study was led by Marija Drndić, an associate professor in the Department of Physics and Astronomy in the School of Arts and Sciences, along with graduate students Kimberly Venta and Matthew Puster and post-doctoral researchers Gabriel Shemer, Julio A. Rodriguez-Manzo and Adrian Balan. They collaborated with assistant professor Jacob K. Rosenstein of Brown University and professor Kenneth L. Shepardof Columbia University.
Their results were published in the journal ACS Nano.
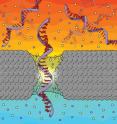
In this technique, known as DNA translocation measurements, strands of DNA in a salt solution are driven through an opening in a membrane by an applied electric field. As each base of the strand passes through the pore, it blocks some ions from passing through at the same time; amplifiers attached to the nanopore chip can register the resulting drop in electrical current. Because each base has a different size, researchers hope to use this data to infer the order of the bases as the strand passes through. The differences in base sizes are so small, however, that the proportions of both the nanopores and membranes need to be close those of the DNA strands themselves -- a major challenge.
The nanopore devices closest to being a commercially viable option for sequencing are made out of protein pores and lipid bilayers. Such protein pores have desirable proportions, but the lipid bilayer membranes in which they are inserted are akin to a film of soap, which leaves much to be desired in terms of durability and robustness.
Solid-state nanopore devices, which are made of thin solid-state membranes, offer advantages over their biological counterparts -- they can be more easily shipped and integrated with other electronics -- but the basic demonstrations of proof-of-principle sensitivity to different DNA bases have been slower.
"While biological nanopores have shown the ability to resolve single nucleotides, solid-state alternatives have lagged due to two challenges of actually manufacturing the right-sized pores and achieving high-signal, low-noise and high-bandwidth measurements," Drndić said. "We're attacking those two challenges here."
Because the mechanism by which the nanopore differentiate between one type of base and another is by the amount of the pore's aperture that is blocked, the smaller a pore's diameter, the more accurate it is. For the nanopore to be effective at determining a sequence of bases, its diameter must approach the diameter of the DNA and its thickness must approach that of the space between one base and the next, or about 0.3 nanometers.
To get solid-state nanopores and membranes in these tiny proportions, researchers, including Drndić's group, are investigating cutting-edge materials, such as graphene. A single layer of carbon atoms in a hexagonal lattice, graphene membranes can be made a little as about 0.5 nanometers thick but have their own disadvantages to be addressed. For example, the material itself is hydrophobic, making it more difficult to pass strands of DNA through them.
In this experiment, Drndić and her colleagues worked with a different material -- silicon nitride -- rather than attempting to craft single-atom-thick graphene membranes for nanopores. Treated silicon nitride is hydrophilic and has readily allowed DNA translocations, as measured by many other researchers during the last decade. And while their membrane is thicker, about 5 nanometers, silicon nitride pores can also approach graphene in terms of thinness due to the way they are manufactured.
"The way we make the nanopores in silicon nitride makes them taper off, so that the effective thickness is about a third of the rest of the membrane," Drndić said.
Drndić and her colleagues tested their silicon nitride nanopore on homopolymers, or single strands of DNA with sequences that consist of only one base repeated several times. The researchers were able to make distinct measurements for three of the four bases: adenine, cytosine and thymine. They did not attempt to measure guanine as homopolymers made with that base bind back on themselves, making it more difficult to pass them through the nanopores.
"We show that these small pores are sensitive to the base content," Drndić said, "and we saw these results in pores with diameters between 1 and 2 nanometers, which is actually encouraging because it suggests some manufacturing variability may be okay."
Source: University of Pennsylvania
Other sources
- Advance in nanotech gene sequencing techniquefrom Science DailyMon, 20 May 2013, 18:00:12 UTC
- Advance in nanotech gene sequencing techniquefrom PhysorgMon, 20 May 2013, 17:00:39 UTC