In CRISPR genome editing, Cpf1, proved its marked specificity and produced a mutant mouse
Related images
(click to enlarge)
As a new tool in CRISPR genome editing, Cpf1 has sparked an explosion of interest for its attributes that differ from Cas9: It requires only a single RNA that CRISPR RNA assembly is simpler; its staggered cleavage patterns may facilitate substituting existing DNA with desired sequences; and it recognizes thymidine-rich DNA sequences, which has been less explored than the guanosine-rich sequences recognized by Cas9. In sum, Cpf1 is expected to broaden the scope of CRISPR genome editing target sites with enhanced efficiency. Despite Cpf1's vast potential as a powerful genome editing tool, little has been demonstrated as to how, specifically, the new tool finds its targets. In a series of two papers published online on June 6 in Nature Biotechnology, researchers at the IBS Center for Genome Editing in South Korea showed Cpf1 as a highly specific programmable tool that is suitable for precision genome editing and reported generation of mutant mice using CRISPR-Cpf1. The researchers used two types of Cpf1 family proteins (AsCpf1 and LbCpf) as they are the most efficient among its kind and performed Digenome-seq: A test they devised to identify intended (on-target) and unintended (off-target) sites the researchers wanted Cpf1 to make a cut throughout the genome. Cell-free genomic DNA isolated from human cells was cleaved with preassembled, recombinant Cpf1 RNPs and subjected to whole-genome sequencing. Uniform alignments of sequence reads, corresponding to on-target and off-target in vitro cleavage sites, were computationally identified.
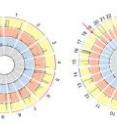
The genome-wide analyses showed that Cpf1 was highly specific, showing fewer off-target cleavage sites (6 for LbCpf1 and 12 for AsCpf1), compared to Cas9, cleaving at >90 sites in the human genome (Fig. 1a). Specifically, at most of the in vitro cleavage sites, indels (representing off-target effects) were below 0.1%, much lower than those at corresponding on-target sites, suggesting that the two Cpf1 proteins had virtually no off-target effects. "Notably, both LbCpf1 and AsCpf1 targeted to a certain site cleaved only the on-target site in the entire human genome" said KIM Daesik, one of first authors of the study (Fig. 1b).
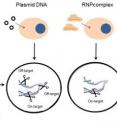
"To reduce off-target effects, we introduced preassembled Cpf1 RNPs into cells, assuming that Cpf1 RNPs, similar to Cas9 RNPs, would cleave target sites immediately after transfection and would be degraded rapidly by endogenous enzymes that break the proteins, thereby reducing off-target effects without sacrificing on-target effects." said KIM Jin-Soo, the corresponding author of the two papers and Director of the IBS Center. Indeed, Cpf1 RNPs did not induce any indels high enough to produce mutations at off-target sites (Fig. 2).
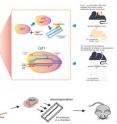
Demonstrating Cpf1's noticeable specificity, another research team from the same IBS Center succeeded in bringing Cpf1 RNP-mediated mutations into mouse embryos: The researchers targeted Foxn1 (a transcription factor that regulates the immune system, including the growth of skin hairs), as well as Tyrosinase (an enzyme that catalyzes the production of melanin, a natural pigment that determines the color of skin). HUR K Junho, first author of the study said, "The data showed that Cpf1 RNP's delivery to mouse embryos resulted in knocking out the intended genetic functions with high mutation frequencies, 64% and 33% respectively. We transplanted the mutant mouse embryos into surrogate mothers and obtained mice with targeted mutations. The mutations resulted in hairless and white-haired mouse respectively (Fig. 3a)." Hur added, "To investigate whether Cpf1 had off-target effects, we performed whole genome sequencing using genomic DNA isolated from one Foxn1 mutant mouse and its wild-type sibling. The sequence analysis showed that no off-target mutation occurred. Targeted deep sequencing of other mutants also showed no off-target mutation"
In the study, the researchers used electrical pulse to permeate Cpf1 RNPs into up to 50 mouse embryos simultaneously (Fig. 3b). This new electroporation delivered the genome-editing molecular for generating mutant animals. Compared to conventional microinjection, the electroporation method was easy to carry out, fast and scalable.
Adding Cpf1 to its toolbox, CRISPR editing technology can build more diverse mouse models. KIM Jin-soo, Director of the IBS Center for Genome Editing and corresponding author of the two studies commented, "Since the two studies have proved the superior specificity of Cpf1, this new nuclease will be more widely used for precise genome editing that does not produce any unintended mutations. CRISPR Cpf1's applications will not be limited, but remain open, starting from therapeutic treatments such as nontoxic cancer drugs, stem cell treatments to highly value-added livestock and agricultural products."
Source: Institute for Basic Science
Other sources
- In CRISPR genome editing, Cpf1, proved its marked specificity and produced a mutant mousefrom Biology News NetMon, 6 Jun 2016, 22:20:58 UTC
- New tool in CRISPR genome editing, Cpf1, proved its marked specificity, produced a targeted mutant mousefrom Science DailyMon, 6 Jun 2016, 18:00:59 UTC
- In CRISPR genome editing, Cpf1, proved its marked specificity and produced a mutant mousefrom PhysorgMon, 6 Jun 2016, 16:01:04 UTC